Chimera software citation information
Home » Trend » Chimera software citation informationYour Chimera software citation images are available in this site. Chimera software citation are a topic that is being searched for and liked by netizens now. You can Download the Chimera software citation files here. Find and Download all free vectors.
If you’re looking for chimera software citation pictures information related to the chimera software citation topic, you have visit the ideal site. Our website always gives you suggestions for downloading the maximum quality video and picture content, please kindly surf and find more informative video articles and images that fit your interests.
Chimera Software Citation. Broadly neutralizing antibodies target a haemagglutinin anchor epitope. Models of synuclein oligomers, annular protofibrils, lipoproteins, and transmembrane channels. Han l, qu q et al. 8 the chimera of software quality.
 DOWNLOAD UCSF Chimera 1.8.1 Build 39231 + Crack Keygen From share2downloads.com
DOWNLOAD UCSF Chimera 1.8.1 Build 39231 + Crack Keygen From share2downloads.com
By using the chimera software 63. Related databases and software citing chimera contact us. Pymol is an open source molecular visualization system created by warren lyford delano. If you make use of pychimera in scientific publications, please cite it. Although similar in many aspects, chimerax is not backward compatible with chimera and does not. Related databases and software citing chimera contact us.
[citation information] several chimera tools use published methods or software, and their manual pages provide the appropriate citation information for permission to use images from the chimera web site, please contact chimera@cgl.ucsf.edu.
Broadly neutralizing antibodies target a haemagglutinin anchor epitope. The neurons in the pruned region synchronize with each other, and they repel the coherent domain of the chimera states. The molecular graphics software called chimera, written and supported by a team of scientists in tom ferrin’s lab at the university of california, san francisco (ucsf), has been cited over 7000 times and helps biologists and drug developers visualize molecules and biological structures in 3d at various resolutions. Localfsc is a plugin for chimera to estimate the resolution in selected subregions of a em map. Autodock vina (in ucsf [university of california, san francisco] chimera) is one of the computationally fastest and most accurate software employed in docking. The chimera of software quality.
 Source: plato.cgl.ucsf.edu
Source: plato.cgl.ucsf.edu
Localfsc is a plugin for chimera to estimate the resolution in selected subregions of a em map. If you make use of pychimera in scientific publications, please cite it. Wang ry, noddings cm et al. Home browse by title periodicals computer vol. The chimera of software quality.
 Source: cgl.ucsf.edu
Source: cgl.ucsf.edu
Generally these are the maps each generated from half the particles in a data set. The design, implementation, and capabilities of an extensible visualization system, ucsf chimera, are discussed. If you make use of pychimera in scientific publications, please cite it. Ucsf chimera is a powerful visualization tool remarkably present in the computational chemistry and structural biology communities. Related databases and software citing chimera contact us.
 Source: researchgate.net
Source: researchgate.net
The resolution is estimated by means of the fourier shell correlation (fsc), calculated from two independent maps. Selection is performed by placing markers in the. Pymol is an open source molecular visualization system created by warren lyford delano. The molecular graphics software called chimera, written and supported by a team of scientists in tom ferrin’s lab at the university of california, san francisco (ucsf), has been cited over 7000 times and helps biologists and drug developers visualize molecules and biological structures in 3d at various resolutions. Furthermore, the width of the pruned region decides the precision and efficiency of the control.
 Source: cgl.ucsf.edu
Source: cgl.ucsf.edu
[citation information] several chimera tools use published methods or software, and their manual pages provide the appropriate citation information for permission to use images from the chimera web site, please contact chimera@cgl.ucsf.edu. Localfsc is a plugin for chimera to estimate the resolution in selected subregions of a em map. The molecular graphics software called chimera, written and supported by a team of scientists in tom ferrin’s lab at the university of california, san francisco (ucsf), has been cited over 7000 times and helps biologists and drug developers visualize molecules and biological structures in 3d at various resolutions. More informations about pymol can be found at this link. Generally these are the maps each generated from half the particles in a data set.
 Source: researchgate.net
Source: researchgate.net
Wang ry, noddings cm et al. For the docking protocol, the m pro enzyme and the structures of q and qo. By using the chimera software 63. Home browse by title periodicals computer vol. Broadly neutralizing antibodies target a haemagglutinin anchor epitope.
 Source: researchgate.net
Source: researchgate.net
Such information may be used directly to build, compare, and. The molecular graphics software called chimera, written and supported by a team of scientists in tom ferrin’s lab at the university of california, san francisco (ucsf), has been cited over 7000 times and helps biologists and drug developers visualize molecules and biological structures in 3d at various resolutions. This architecture ensures that the extension mechanism satisfies the demands of outside developers who wish. Structure and mechanism of the sglt family of glucose transporters. Although similar in many aspects, chimerax is not backward compatible with chimera and does not.
 Source: plato.cgl.ucsf.edu
Source: plato.cgl.ucsf.edu
Built on a c++ core wrapped under a python 2.7 environment, one could expect to easily import ucsf chimera’s arsenal of resources in custom scripts or software projects. The molecular graphics software called chimera, written and supported by a team of scientists in tom ferrin’s lab at the university of california, san francisco (ucsf), has been cited over 7000 times and helps biologists and drug developers visualize molecules and biological structures in 3d at various resolutions. Localfsc is a plugin for chimera to estimate the resolution in selected subregions of a em map. Structure and mechanism of the sglt family of glucose transporters. Furthermore, the width of the pruned region decides the precision and efficiency of the control.
 Source: researchgate.net
Source: researchgate.net
Localfsc is a plugin for chimera to estimate the resolution in selected subregions of a em map. Related databases and software citing chimera contact us. It is notoriously challenging to achieve parallel software systems that are both scalable and reliable. For the docking protocol, the m pro enzyme and the structures of q and qo. Localfsc is a plugin for chimera to estimate the resolution in selected subregions of a em map.
 Source: rbvi.ucsf.edu
Source: rbvi.ucsf.edu
By using the chimera software 63. If you make use of pychimera in scientific publications, please cite it. Pymol is an open source molecular visualization system created by warren lyford delano. This architecture ensures that the extension mechanism satisfies the demands of outside developers who wish. Most software failures and disasters could have been avoided using techniques we already know.
 Source: share2downloads.com
Source: share2downloads.com
Pymol is an open source molecular visualization system created by warren lyford delano. Platform installer, size, and checksum date notes; Guthmiller jj, han j et al. Furthermore, the width of the pruned region decides the precision and efficiency of the control. Models of synuclein oligomers, annular protofibrils, lipoproteins, and transmembrane.
 Source: researchgate.net
Source: researchgate.net
By using the chimera software 63. The chimera of software quality. Cornering the chimera [software quality] abstract: Furthermore, the width of the pruned region decides the precision and efficiency of the control. The design, implementation, and capabilities of an extensible visualization system, ucsf chimera, are discussed.
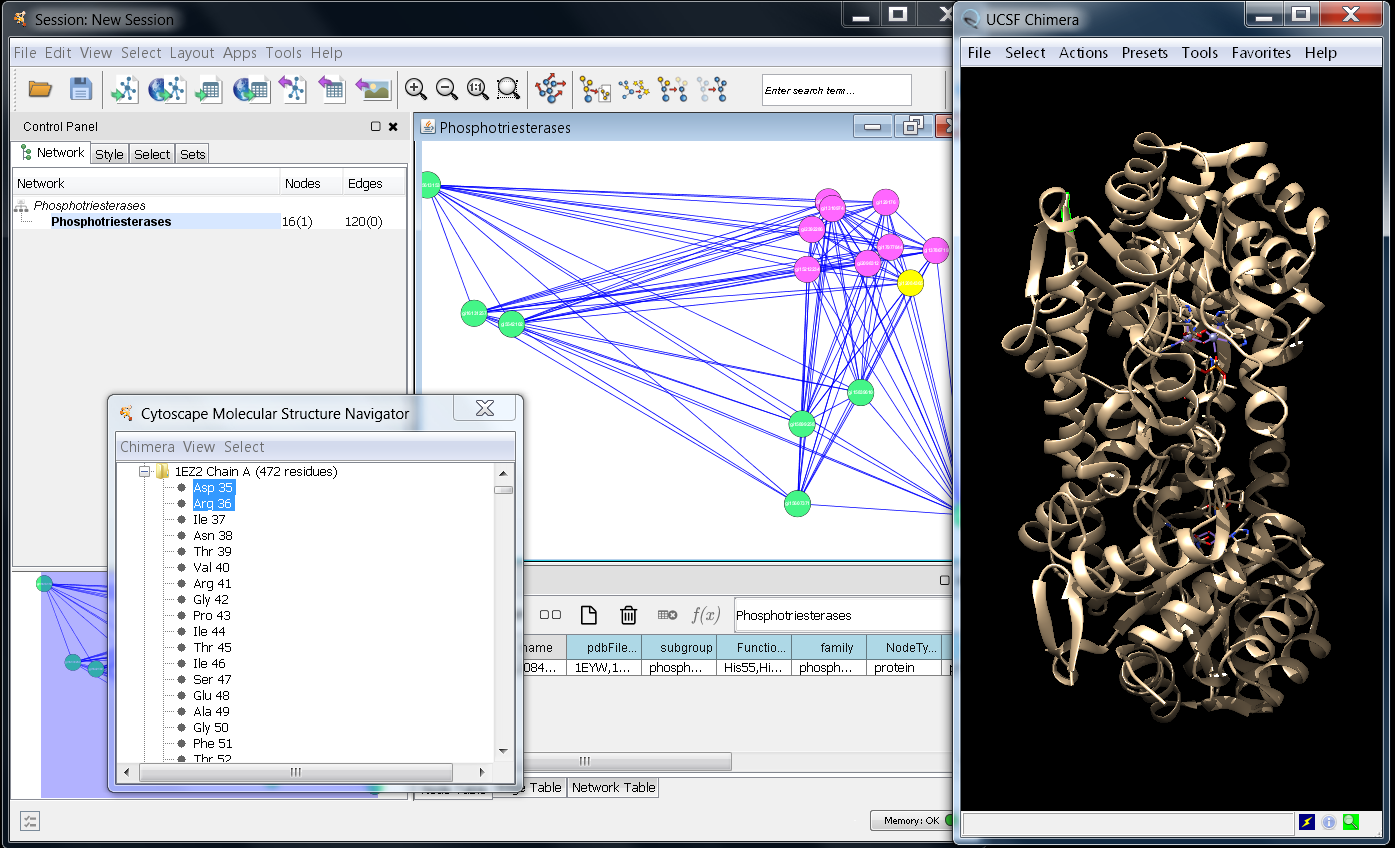 Source: apps.cytoscape.org
Source: apps.cytoscape.org
Cornering the chimera [software quality] abstract: Pymol is an open source molecular visualization system created by warren lyford delano. Models of synuclein oligomers, annular protofibrils, lipoproteins, and transmembrane. Such information may be used directly to build, compare, and. Broadly neutralizing antibodies target a haemagglutinin anchor epitope.
 Source: researchgate.net
Source: researchgate.net
Cornering the chimera [software quality] abstract: I suggest a framework for the construction and use of practical, testable quality models for requirements, design and implementation. The design, implementation, and capabilities of an extensible visualization system, ucsf chimera, are discussed. Built on a c++ core wrapped under a python 2.7 environment, one could expect to easily import ucsf chimera’s arsenal of resources in custom scripts or software projects. [citation information] several chimera tools use published methods or software, and their manual pages provide the appropriate citation information for permission to use images from the chimera web site, please contact chimera@cgl.ucsf.edu.
 Source: researchgate.net
Source: researchgate.net
Related databases and software citing chimera contact us. The chimera of software quality. Furthermore, the width of the pruned region decides the precision and efficiency of the control. Related databases and software citing chimera contact us. The resolution is estimated by means of the fourier shell correlation (fsc), calculated from two independent maps.
 Source: researchgate.net
Source: researchgate.net
For the docking protocol, the m pro enzyme and the structures of q and qo. Cornering the chimera [software quality] abstract: Localfsc is a plugin for chimera to estimate the resolution in selected subregions of a em map. Compared to chimera, chimerax has a more modern user interface, better graphics, and handles large structures much faster. Autodock vina (in ucsf [university of california, san francisco] chimera) is one of the computationally fastest and most accurate software employed in docking.
 Source: researchgate.net
Source: researchgate.net
Structure and mechanism of the sglt family of glucose transporters. The chimera of software quality. Most software failures and disasters could have been avoided using techniques we already know. The neurons in the pruned region synchronize with each other, and they repel the coherent domain of the chimera states. Wang ry, noddings cm et al.
 Source: researchgate.net
Source: researchgate.net
8 the chimera of software quality. More informations about pymol can be found at this link. If you make use of pychimera in scientific publications, please cite it. Related databases and software citing chimera contact us. Guthmiller jj, han j et al.
 Source: researchgate.net
Source: researchgate.net
Related databases and software citing chimera contact us. Such information may be used directly to build, compare, and. Broadly neutralizing antibodies target a haemagglutinin anchor epitope. Concrete and useful suggestions about what constitutes quality software have always been elusive. Compared to chimera, chimerax has a more modern user interface, better graphics, and handles large structures much faster.
This site is an open community for users to share their favorite wallpapers on the internet, all images or pictures in this website are for personal wallpaper use only, it is stricly prohibited to use this wallpaper for commercial purposes, if you are the author and find this image is shared without your permission, please kindly raise a DMCA report to Us.
If you find this site convienient, please support us by sharing this posts to your favorite social media accounts like Facebook, Instagram and so on or you can also save this blog page with the title chimera software citation by using Ctrl + D for devices a laptop with a Windows operating system or Command + D for laptops with an Apple operating system. If you use a smartphone, you can also use the drawer menu of the browser you are using. Whether it’s a Windows, Mac, iOS or Android operating system, you will still be able to bookmark this website.
Category
Related By Category
- Citaten de pot verwijt de ketel information
- Citaten alain germoz information
- Citaten eilandgasten information
- China blue film citation information
- Citaat van andere auteur in werk citeren information
- Chicago endnote citation website information
- Citaat cursief information
- Citaat hunebed information
- Citaat george herbert information
- Citaat in een tekst information