Cipres science gateway citation information
Home » Trend » Cipres science gateway citation informationYour Cipres science gateway citation images are ready in this website. Cipres science gateway citation are a topic that is being searched for and liked by netizens now. You can Download the Cipres science gateway citation files here. Download all royalty-free vectors.
If you’re looking for cipres science gateway citation pictures information connected with to the cipres science gateway citation topic, you have visit the ideal blog. Our website always provides you with suggestions for seeking the highest quality video and image content, please kindly hunt and find more enlightening video articles and graphics that fit your interests.
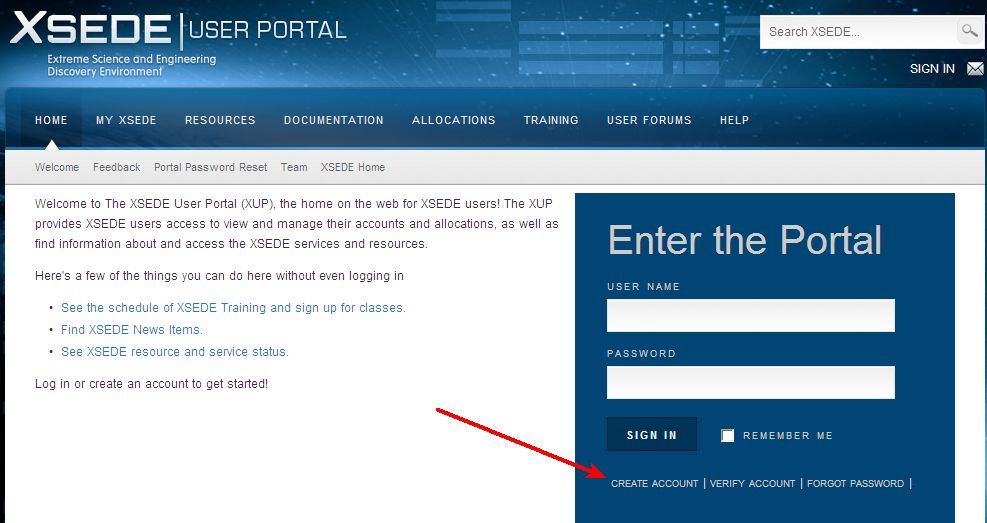
Cipres Science Gateway Citation. The cipres science gateway is a community web application that provides public access to a set of parallel tree inference and multiple sequence alignment codes run on large computational resources. But i am curious, the documentation says phased is for when linkage=1. Also, i cant find documentation for the revert_convert=1 command. Cipres stands for cyberinfrastructure for phylogenetic research, and xsede stands for extreme.
 SDSC CIPRES Gateway Help Define New “Tree From ucsdnews.ucsd.edu
SDSC CIPRES Gateway Help Define New “Tree From ucsdnews.ucsd.edu
The constraint s option allows you to specify complicated prior probabilities on trees (constraints are discussed more fully in help constraint). It is designed to provide all researchers with access to nsf xsede �s large computational resources through a simple browser interface. This paper describes development of the cipres science gateway, a web portal designed to provide researchers with transparent access to the fastest available community codes for inference of phylogenetic relationships, and implementation of these codes on scalable computational resources. To cipres science gateway users. • access to most/all native command line options for several codes. To cipres science gateway users.
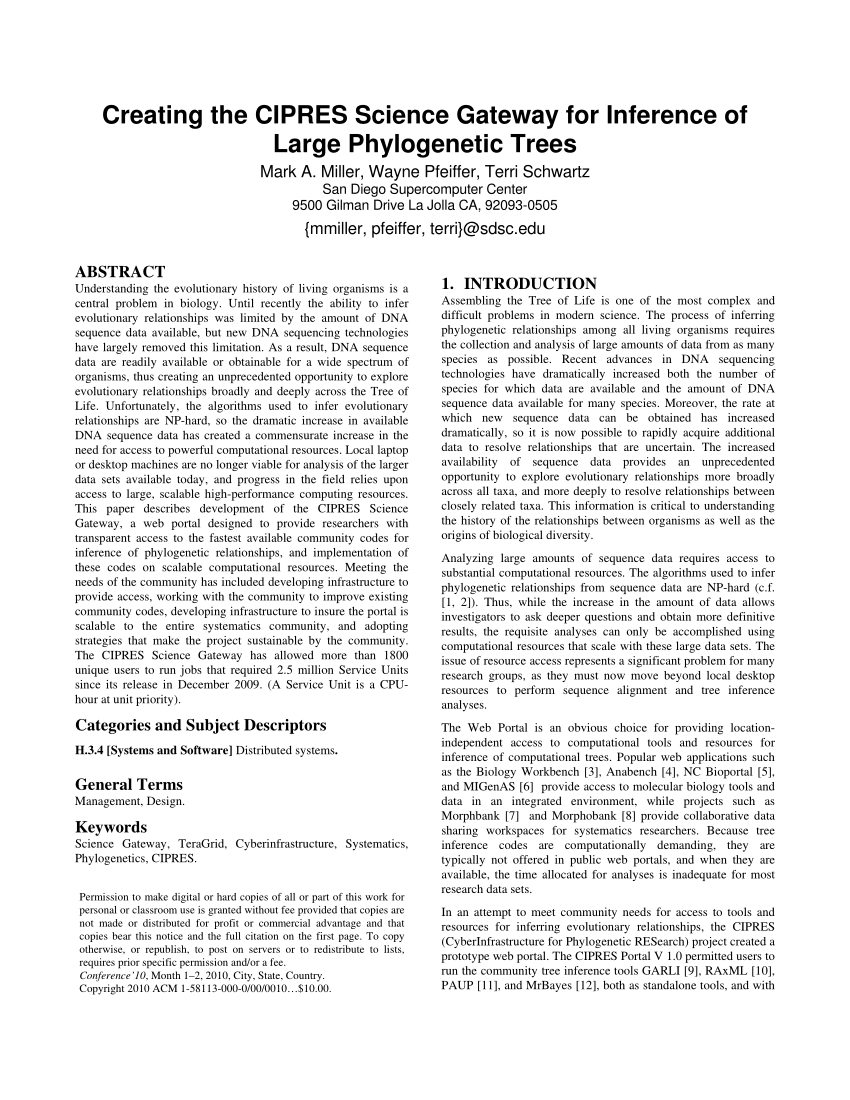
This paper describes development of the cipres science gateway, a web portal designed to provide researchers with transparent access to the fastest available community codes for inference of phylogenetic relationships, and implementation of these codes on scalable computational resources.
This paper describes development of the cipres science gateway, a web portal designed to provide researchers with transparent access to the fastest available community codes for inference of phylogenetic relationships, and implementation of these codes on scalable computational resources. Cipres stands for cyberinfrastructure for phylogenetic research, and xsede stands for extreme. It is designed to provide all researchers with access to nsf xsede �s large computational resources through a simple browser interface. Also, i cant find documentation for the revert_convert=1 command. Creating the cipres science gateway for inference of large phylogenetic trees. A., pfeiffer, w., & schwartz, t.
 Source: researchgate.net
Source: researchgate.net
Science portals & science gateways. This can be answered using the mb manual. I am not sure if that is right. Hi there, thanks for the question. I have the interface just about ready so you can urn this command line.
 Source: phylo.org
Source: phylo.org
Miller, m.a., pfeiffer, w., and schwartz, t. Hi there, thanks for the question. I have the interface just about ready so you can urn this command line. (2010) creating the cipres science gateway for inference of large phylogenetic trees in proceedings of the gateway computing environments workshop (gce), 14 nov. Understanding the evolutionary history of living organisms is a central problem in biology.
![[PDF] Creating the CIPRES Science Gateway for inference of [PDF] Creating the CIPRES Science Gateway for inference of](https://d3i71xaburhd42.cloudfront.net/e27acaf97f5b2eae4257bb5d8278fbe0e6405c39/5-Figure2-1.png) Source: semanticscholar.org
Source: semanticscholar.org
3.3 is a public resource for inference of large phylogenetic trees. Hi there, thanks for the question. The cipres science gateway v. I have the interface just about ready so you can urn this command line. This forum is for sharing questions and issues in using the cipres resource.
 Source: ucsdnews.ucsd.edu
Source: ucsdnews.ucsd.edu
• access to most/all native command line options for several codes. This can be answered using the mb manual. This forum is for sharing questions and issues in using the cipres resource. But you used it with linkage=0. Also, i cant find documentation for the revert_convert=1 command.
 Source: blog.csdn.net
Source: blog.csdn.net
A new web resource developed at the san diego supercomputer. The new nsf award, which follows an earlier nsf grant that ran from 2003 to 2008, is for the cipres science gateway, a web site that allows researchers to explore evolutionary relationships between species using supercomputers provided by the nsf xsede project. • access to most/all native command line options for several codes. Miller, m.a., pfeiffer, w., and schwartz, t. In 2010 gateway computing environments workshop (gce) (pp.
 Source: researchgate.net
Source: researchgate.net
Cipres home » about » cite_us if you use the resources available from the cipres science gateway to complete published work, please cite us as follows: Miller, m.a., pfeiffer, w., and schwartz, t. Until recently the ability to infer evolutionary relationships was limited by the amount of dna sequence data available, but new dna sequencing technologies have largely removed this. It is designed to provide all researchers with access to nsf xsede �s large computational resources through a simple browser interface. The cipres science gateway v.
 Source: researchgate.net
Source: researchgate.net
Cipres science gateway clarifies branches in evolution�s �tree of life�. 3.3 is a public resource for inference of large phylogenetic trees. But i am curious, the documentation says phased is for when linkage=1. The functionalities and concepts were created by a team that included tandy warnow, bernard moret, mark holder, paul hoover, lucie chan, terri schwartz, rutger vos, mark miller, peter midford, andres varon, and wayne. The constraint s option allows you to specify complicated prior probabilities on trees (constraints are discussed more fully in help constraint).
 Source: researchgate.net
Source: researchgate.net
Creating the cipres science gateway for inference of large phylogenetic trees abstract: The cipres science gateway is a community web application that provides public access to a set of parallel tree inference and multiple sequence alignment codes run on large computational resources. To cipres science gateway users. Hi there, thanks for the question. You can also access these same capabilities programatically with the cipres rest api.
 Source: researchgate.net
Source: researchgate.net
• access to most/all native command line options for several codes. The cipres science gateway was designed to allow users to analyze large sequence data sets using community codes on significant computational resources. But i am curious, the documentation says phased is for when linkage=1. To cipres science gateway users. Science portals & science gateways.
 Source: pdfcoffee.com
Source: pdfcoffee.com
This paper describes development of the cipres science gateway, a web portal designed to provide researchers with transparent access to the fastest available community codes for. Cipres science gateway clarifies branches in evolution�s �tree of life�. The cipres science gateway v. I have the interface just about ready so you can urn this command line. This paper describes development of the cipres science gateway, a web portal designed to provide researchers with transparent access to the fastest available community codes for inference of phylogenetic relationships, and implementation of these codes on scalable computational resources.
 Source: researchgate.net
Source: researchgate.net
The cipres science gateway is a community web application that provides public access to a set of parallel tree inference and multiple sequence alignment codes run on large computational resources. (2010) creating the cipres science gateway for inference of large phylogenetic trees in proceedings of the gateway computing environments workshop (gce), 14 nov. This can be answered using the mb manual. Creating the cipres science gateway for inference of large phylogenetic trees. It is designed to provide all researchers with access to nsf xsede �s large computational resources through a simple browser interface.
![[PDF] Creating the CIPRES Science Gateway for inference of [PDF] Creating the CIPRES Science Gateway for inference of](https://d3i71xaburhd42.cloudfront.net/e27acaf97f5b2eae4257bb5d8278fbe0e6405c39/3-Figure1-1.png) Source: semanticscholar.org
Source: semanticscholar.org
But you used it with linkage=0. These resources are made available at no charge to users by the nsf extreme science and engineering discovery environment (xsede) project. This paper describes development of the cipres science gateway, a web portal designed to provide researchers with transparent access to the fastest available community codes for inference of phylogenetic relationships, and implementation of these codes on scalable computational resources. The cipres science gateway v. But you used it with linkage=0.
 Source: researchgate.net
Source: researchgate.net
This forum is for sharing questions and issues in using the cipres resource. 3.3 is a public resource for inference of large phylogenetic trees. The functionalities and concepts were created by a team that included tandy warnow, bernard moret, mark holder, paul hoover, lucie chan, terri schwartz, rutger vos, mark miller, peter midford, andres varon, and wayne. (2010) creating the cipres science gateway for inference of large phylogenetic trees in proceedings of the gateway computing environments workshop (gce), 14 nov. To cipres science gateway users.
 Source: frontiersin.org
Source: frontiersin.org
I am not sure if that is right. You can also access these same capabilities programatically with the cipres rest api. These resources are made available at no charge to users by the nsf extreme science and engineering discovery environment (xsede) project. This paper describes development of the cipres science gateway, a web portal designed to provide researchers with transparent access to the fastest available community codes for. Until recently the ability to infer evolutionary relationships was limited by the amount of dna sequence data available, but new dna sequencing technologies have largely removed this.
 Source: youtube.com
Source: youtube.com
The functionalities and concepts were created by a team that included tandy warnow, bernard moret, mark holder, paul hoover, lucie chan, terri schwartz, rutger vos, mark miller, peter midford, andres varon, and wayne. The cipres science gateway was designed to allow users to analyze large sequence data sets using community codes on significant computational resources. You can also access these same capabilities programatically with the cipres rest api. The cipres science gateway is a community web application that provides public access to a set of parallel tree inference and multiple sequence alignment codes run on large computational resources. Cipres science gateway clarifies branches in evolution�s �tree of life�.
![[PDF] Creating the CIPRES Science Gateway for inference of [PDF] Creating the CIPRES Science Gateway for inference of](https://d3i71xaburhd42.cloudfront.net/e27acaf97f5b2eae4257bb5d8278fbe0e6405c39/5-Table1-1.png) Source: semanticscholar.org
Source: semanticscholar.org
The new nsf award, which follows an earlier nsf grant that ran from 2003 to 2008, is for the cipres science gateway, a web site that allows researchers to explore evolutionary relationships between species using supercomputers provided by the nsf xsede project. To cipres science gateway users. These resources are made available at no charge to users by the nsf extreme science and engineering discovery environment (xsede) project. This paper describes development of the cipres science gateway, a web portal designed to provide researchers with transparent access to the fastest available community codes for inference of phylogenetic relationships, and implementation of these codes on scalable computational resources. (2010) creating the cipres science gateway for inference of large phylogenetic trees in proceedings of the gateway computing environments workshop (gce), 14 nov.
 Source: nap.edu
Source: nap.edu
Miller, m.a., pfeiffer, w., and schwartz, t. (2010) creating the cipres science gateway for inference of large phylogenetic trees in proceedings of the gateway computing environments workshop (gce), 14 nov. The new nsf award, which follows an earlier nsf grant that ran from 2003 to 2008, is for the cipres science gateway, a web site that allows researchers to explore evolutionary relationships between species using supercomputers provided by the nsf xsede project. You can also access these same capabilities programatically with the cipres rest api. • access to most/all native command line options for several codes.
 Source: researchgate.net
Source: researchgate.net
The constraint s option allows you to specify complicated prior probabilities on trees (constraints are discussed more fully in help constraint). A., pfeiffer, w., & schwartz, t. Miller, m.a., pfeiffer, w., and schwartz, t. But you used it with linkage=0. The constraint s option allows you to specify complicated prior probabilities on trees (constraints are discussed more fully in help constraint).
This site is an open community for users to do submittion their favorite wallpapers on the internet, all images or pictures in this website are for personal wallpaper use only, it is stricly prohibited to use this wallpaper for commercial purposes, if you are the author and find this image is shared without your permission, please kindly raise a DMCA report to Us.
If you find this site convienient, please support us by sharing this posts to your preference social media accounts like Facebook, Instagram and so on or you can also bookmark this blog page with the title cipres science gateway citation by using Ctrl + D for devices a laptop with a Windows operating system or Command + D for laptops with an Apple operating system. If you use a smartphone, you can also use the drawer menu of the browser you are using. Whether it’s a Windows, Mac, iOS or Android operating system, you will still be able to bookmark this website.
Category
Related By Category
- Citaten de pot verwijt de ketel information
- Citaten alain germoz information
- Citaten eilandgasten information
- China blue film citation information
- Citaat van andere auteur in werk citeren information
- Chicago endnote citation website information
- Citaat cursief information
- Citaat hunebed information
- Citaat george herbert information
- Citaat in een tekst information